Division of Molecular Biosciences
Department of Life Sciences
Faculty of Natural Sciences

|
|
|
Research OverviewContents
Biographical background (Professor Sternberg)
|
|||||||
The current objectives of the Structural Bioinformatics Group are:
The group's web page (http://www.sbg.bio.ic.ac.uk) provides access to web servers for several areas of protein modelling including protein structure prediction (3D-PSSM / PHYRE), protein-protein docking (3D-GARDEN) and protein function prediction (CONFUNC). Of particular note is the use by the community of the programs 3D-PSSM and Phyre. The 3D-PSSM paper has over 1282 citations (ISI Web of Science) and the server web page has had over 250,000 visits. The more recent program Phyre has approximately 5000 requests per month. Recent work has extended these concepts to study biology networks and to identify novel drugs.
Professor Sternberg's group interacts closely with the groups of Professor Michael Stumpf (Theoretical Systems Biology Group) and Professor Stephen Muggleton (in the use of advanced machine learning) at Imperial. In addition, there are collaborations with members of the Centre for Integrative Systems Biology at Imperial College (CISBIC) and the Institute of Systems and Synthetic Biology.
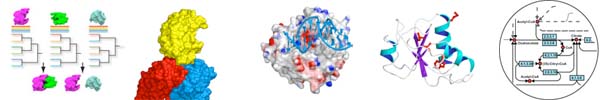
The Protein Homology/Analogy Recognition Engine (Phyre) is a user-friendly and widely used server for automated 3D protein modelling, averaging 150 submissions per day. A user may paste their protein sequence into the browser and will receive a notification email when modelling is complete (usually within approx. 30mins-1hr). This email contains the top scoring 3D model of the user's protein and a link to a web page containing a wide range of predicted structural features, including secondary structure, disorder, potential binding sites, SCOP fold and superfamily annotations, and 3D models with confidence estimates for the top 10 matching potential homologous structures. Phyre is capable of detecting remote homology to known structures significantly beyond the range of the popular PSI-Blast. By using advanced profile-profile matching techniques, loop modelling and sidechain placement algorithms, accurate full-atom models can be built based on homology to known protein structures with sequence identities <15%.
A new and more powerful version of Phyre (Phyre de novo) has recently been tested in the international CASP8 structure prediction competition and was ranked amongst the best performers. This new system contains advanced homology detection features, multi-domain modelling and ab initio components. We hope to release this new version of the server in the autumn of 2009. The Phyre server is available at http://www.sbg.bio.ic.ac.uk/phyre/.
Poing is a model for protein folding based upon Langevin dynamics, with the primary aim of predicting the structure of a protein from its sequence, without making use of template structures with homologous sequence. The current focus of this work is upon using general statistical and geometric featu res of known natural protein str uctures to identify good and bad structures from the ensemble generated by the model. It is simple to add features of the cellular environment to the model, which can be used to investigate their effects upon protein folding. This picture shows a model of a protein being synthesized into a crowded cell (grey translucent spheres) by a ribosome (blue, on the right). This work is funded by the BBSRC.
The thousands of sequenced genomes and millions of sequences identified
by metagenomics projects make the prediction of protein function an
important problem. While function prediction can be relatively simple
when sequences share high levels of similarity, it is cases where
sequences only have more remote homologues that current function
prediction methods are ineffective. This has led to the development of
ConFunc, a sequence based protein function method that complement existing
tools by performing well for these more difficult cases. ConFunc identifies
GO annotated sequences present in PSI-BLAST searches and uses these to
identify conserved residues associated with each individual function, which
in turn are used to infer the function of query sequences. ConFunc is
available to the academic community via a webserver at
http://www.sbg.bio.ic.ac.uk/confunc/.
Macromolecular docking problems involve predicting the molecular geometry of the
complex formed when two macromolecules associate. 3DGarden is an integrated software
suite for performing protein-protein and protein-polynucleotide docking. For
any pair of biomolecules structures specified by the user, 3DGarden's primary
function is to generate an ensemble of putative complexed structures and rank them.
The highest-ranking candidates constitute predictions for the structure of the complex.
3DGarden cannot be used to decide whether or not a particular pair of biomolecules
interacts. Complexes of protein and nucleic acid chains can also be specified as
individual interactors for docking purposes.3D-Garden is available to the academic
community via a webserver at
http://www.sbg.bio.ic.ac.uk/3dgarden/.
Understanding protein interactions has broad implications for the mechanism of recognition,
protein design, and assigning putative functions to uncharacterised proteins. Studying
protein flexibility is a key component in the challenge of describing protein interactions.
We have characterised the observed conformational change for a set of 20 proteins that
undergo large conformational change upon association (> 2Å Ca RMSD) and ask what features
of the motion are successfully reproduced by the normal modes of the system. We demonstrated
that normal modes can be used to identify mobile regions and in some proteins to reproduce
the direction of conformational change. In 35% of the proteins studied a single low
frequency normal mode was found that describes the direction of the observed conformational
change well. Finally, we find that for a set of 138 proteins from a docking benchmark that
the characteristic frequencies of normal modes can be used to predict reliably the extent
of conformational change. This study has implications for the mechanics of protein recognition.
We are applying cutting edge machine learning technology to problems of protein function and structure. Coupling the statistical power of Support Vector Machines (SVMs) with the logical relational learning approach Inductive Logic Programming (ILP), we are learning rules that determine the small molecules that bind to a protein active site.
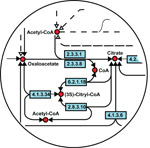
High-throughput technologies have generated large amounts of data on the interactions of macromolecules. However the usage of these interaction data is still limited. The aim of the project is to develop a method to use protein interaction data to infer functions of proteins. The idea is to detect similarities in protein interaction networks which thus helps inferring functions of correlated proteins by function transfer. This project extends previous research in developing a method to compare biological networls PHUNKEE (Cootes et al, 2007).
This project focuses on discovering what effect the massive increase in available protein sequences has had on our ability to identify remote homology. The use of conventional methods shows that we are already facing diminishing returns and the aim of this project is to investe methods to improve homology detection based upon these findings.
The role of modelling in systems biology is to assimilate background and
experimental knowledge and identify new hypotheses which are interesting
and cost-effective to test. Here we use inductive logic programming, a machine
reasoning framework, in a systems biology project investigating the glycomics
behind interactions between pathogenic bacteria and their host. Cell surface
glycans in the form of polysaccharides, glycoproteins and glycolipids are
critical in cell-cell communication and cell signalling. In the context of
host-pathogen interactions, microbial glycans are frequent targets of the
pattern recognition receptors involved in triggering innate immune responses,
and influence the uptake of microbes by host phagocytes. The aim of this part
of the project is to develop a artificial reasoning model that will allow us
to infer the surface glycome from genetic data. Transcriptome and metabolome
data are used to build the models. This project is part of the research studying
host/pathogen interactions in the BBSRC/EPSRC Centre of Intergrative Systems
Biology at Imperial College (CISBIC). The work using inductive logic programming
in collaboration with Professor Stephen Muggleton. This project extends the
methodology develop to model the effects of specfic toxins in metabolic networks.
This project builds upon the paper by Amini, Shrimpton, Muggleton,
and Sternberg (2007) "A general approach for developing system-specific
functions to score
protein-ligand docked complexes using support vector inductive logic
programming" Proteins, 69(4): 823-831. This project will attempt to use
a machine learning algorithm based on logical rules to design new,
original, and more effective pharmaceutical drugs. The project will use
knowledge of chemical synthesis to try and automate initial hit to lead
searching. The overall aims will be to produce a tool that can
contribute to future drug design, and to identify at least one small
molecule with an improved docking affinity to existing drugs with the
same target. This project is in collaboration with Professor Stephen Muggleton.
This technology has been commercialised by a spin-out company Equinox Pharma
Ltd and further details can be found on the companies web pages at http://www.equinoxpharma.com
Analysis and prediction of protein topology - Comparative analyses of the known protein structures
identified fundamental features of their folding topology. The connection between two parallel
strands in the same sheet including the beta-alpha-beta unit was found almost invariably to be right-handed.
Knowledge-based protein structure prediction - Analysis of the packing in beta/alpha and beta/beta proteins identified rules which were used for a knowledge-based approach for structure prediction.
Protein mobility - Analysis of crystallographic B-values (temperature factors) derived from refinement of lysozyme provided insight into protein mobility.
Electrostatic effects in proteins - Modelling electrostatic effects in protein was shown to yield successful blind predictions of experimentally observed pKa shifts.
Novel algorithms for protein sequence alignment - Algorithms were developed to perform multiple alignment of protein sequences and to identify remote homologues using profile scanning.
Relational database of protein structure - A relational database encoding features of protein structure was established. This facilitated rapid analyses to derive novel principles of protein conformation.
Poly-proline helices - A comparative analysis of protein structures showed that the poly-proline helix commonly occur in globular proteins
Application of machine learning to drug design - A logic-based approach to model the structure-activity relationship of small molecules was developed. The approach yields rules which can readily be understood by chemists and can provide predictions of activity (or toxicity) which can be superior to established methods.
DTI MRC (Medical Research Council)
Bang, J. W., Crockford, D. J., Holmes, E., Pazos, F., Sternberg,
M. J. E, Muggleton, S. H. & Nicholson, J. K. (2008). Integrative
top-down system metabolic modeling in experimental disease states
via data-driven Bayesian methods. J Proteome Res 7, 497-503.
Inductive Logic-Based Drug Design
Amin i, A., Muggleton, S. H. Lodhi, H. & Sternberg, M. J.E. (2007).
A Novel Logic-Based Approach for Quantitative Toxicology Prediction. J Chem Inf Model, 2007.47, 998-1006.
Amini, A., Shrimpton, P. J., Muggelton, S. H. & Sternberg, M. J. E. (2007). A general approach
for developing system-specfic scoring functions to score protein-liganddocked complexes using
support vector inductive logic programming. Proteins 2007, 69, 823 - 831
Highlights of previous research
Sternberg, M. J. E. & Thornton, J. M. (1976). On the conformation of proteins: the handedness of β-stand / α-helix / β-strand unit J. Mol. Biol. 105, 367-382.
Sternberg, M. J. E. & Thornton, J. M. (1977). On the conformation of proteins: an analysis of β-pleated sheets. J. Mol. Biol. 110, 285-296.
Sternberg, M. J. E. & Thornton, J. M. (1977). On the conformation of proteins: towards
the prediction of strand arrangements in β-pleated sheets. J. Mol. Biol. 113, 401-418.
Cohen, F. E., Sternberg, M. J. E. & Taylor, W. R. (1980). Analysis and prediction of protein β-sheet structures by a combinatorial approach. Nature, 285, 378-382.
Sternberg, M. J. E., Grace, D. E. & Phillips, D. C. (1979). Dynamic information from protein crystallography. An analysis of temperature factors from refinement of the hen egg-white lysozyme structure. J. Mol. Biol. 130, 231-252.
Artymiuk, P. J., Blake, C. C., Grace, D. E., Oatley, S. J., Phillips, D. C. & Sternberg, M. J. E. (1979). Crystallographic studies of the dynamic properties of lysozyme. Nature, 280, 563-568.
Sternberg, M. J. E., Hayes, F. R., Russell, A. J., Thomas, P. G. & Fersht, A. R. (1987). Prediction of electrostatic effects of engineering of protein charges. Nature, 330, 86-88
Barton, G. J. & Sternberg, M. J. E. (1987). A strategy for the rapid multiple alignment of protein sequences. Confidence levels from tertiary structure comparisons. J. Mol. Biol. 198, 327-337
McGregor, M. J., Islam, S. A. & Sternberg, M. J. E. (1987). Analysis of the relationship between side-chain conformation and secondary structure in globular proteins. J. Mol. Biol. 198, 295-310.
Adzhubei, A. A. & Sternberg, M. J. E. (1993). Left-handed poly-proline II helices commonly occur in globular proteins. J. Mol. Biol. 229, 472-493.
King, R. D., Muggleton, S. H., Srinivasan, A. & Sternberg, M. J. E. (1996). Representing molecular structure activity relationships: the use of atoms and their bond connectivities to predict mutagenicity using inductive logic programming. Proc. Nat. Acad. Sci. USA, 93, 438-442
Funding
![]()
![]()